BioChemiScape is a professional knowledge network for researchers in biochemistry, cell and molecular biology, biomedicine and nanotechnology that promotes open access to multiple types of scientific content
BioChemiScape seeks to open more scientific data to a broader public; to encourage scientists, researchers, key stakeholders and students from the fields of biochemistry, cell and molecular biology, biomedicine and nanotechnology to engage in a multidimensional dialogue concerning the open access to scientific data both in its raw (e.g. data sets) and interpreted (e.g. graphical or summary) forms – including both work-in-progress and real time data as well as final, “published” data. In the project we explore novel paradigms for discovering, querying, linking, aggregating, filtering, enriching, visualizing and publishing scientific data that comes from disparate networks of scientific content owners and providers. Members can interact with the resources, add value by annotating, enriching and visualizing the data; and in turn publish these results.
Overall aim of BioChemiScape
In the BioChemiScape proposal we will explore novel paradigms for discovering, querying, linking, aggregating, filtering, enriching, visualizing and publishing scientific data that comes from disparate networks of scientific content owners and providers in the fields of biochemistry, cell and molecular biology, biomedicine and nanotechnology.
- BioChemiScape seeks to encourage researchers, key stakeholders, students and the public to engage in a multidimensional dialogue concerning the scientific data; in both its raw (e.g. data sets) and interpreted (e.g. graphical or summary) forms – including both work-in-progress and real time data as well as “published” data.
- BioChemiScape provides “data windows” which can be used to retrieve filtered or complete data sets and programmable widgets which can process, perform aggregate operations, display and trigger alerts if certain data conditions are met.
- BioChemiscape provides programmable widgets that can query, arrange and display a selection of articles or synopses based on the member’s follow and filter preferences; and provide links to the raw underlying data of the articles. These programmable data and article widgets may be dragged and dropped on the end member’s virtual dashboard.
- Members can interact with the resources, add value by annotating, enriching and visualizing the data; and in turn publish these results.
- Members can form groups in order to collaboratively publish and aggregate the data in specialized topic nodes.
- BioChemiScape promotes Open Research and Open Access by leveraging Open Data and exposing a developer friendly API and a data widget framework.
Technology
BioChemiScape uses Eviscape, a social knowledge network developed by TESOBE. Eviscape leverages a robust open source stack including Linux, Python, Django, Postgres, RabbitMQ and external NoSQL data stores such as MongoDB – and their associated libraries. This leads to a powerful, dynamic, structured and flexible software infrastructure the components of which are regularly updated by the relevant open source communities. Eviscape itself is supported by broad test coverage and in code documentation facilitating refactoring and agile development practices. The platform can run on commodity hardware and / or in the cloud. Eviscape services are exposed as a RESTful API (XML, JSON etc.) and HTML5 / Javascript interfaces over http / https. This approach lends itself to both rich clients consuming the API and HTML5 powered mobile or desktop apps as well as the main web application. Eviscape provides a data and social / topic graph platform that supports data structuring(XML, JSON), linking, tagging and sharing, social interaction, collaborative processes, multi-party content publication, discovery, semantic filtering, aggregation, automated data harvesting (via RSS), a rich API, a programmable widget infrastructure and external social network integration.
Technical Overview
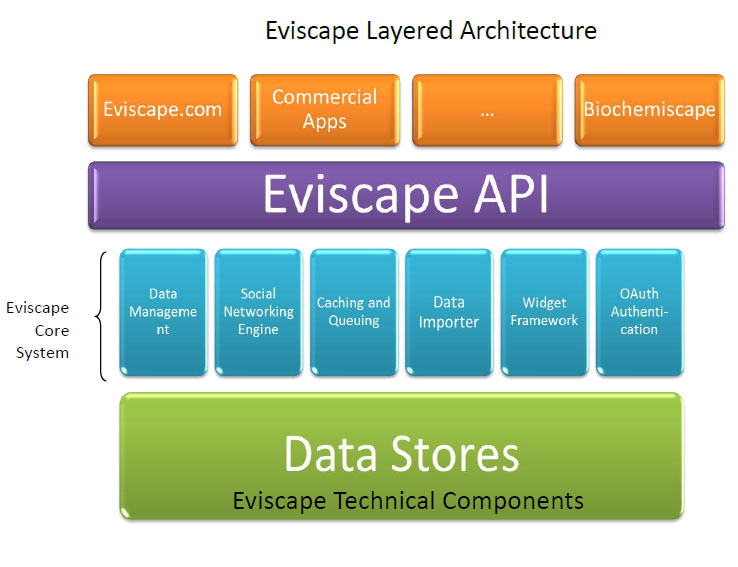
BioChemiScape is based on Eviscape, the architecture of which is diagrammed below:
Four level service diagram:
1) Applications (HTML5 etc.)
2) API (REST)
3) Application code (Python, Django, Javascript)
4) Data stores, database code. (Postgres, PGplsql, PLpythonu)
 e-Commerce Group
e-Commerce Group